Note
Go to the end to download the full example code.
3D translation: image to mask#
This example illustrates coregistration by 3D translation. The moving volume is a 3D magnetization transfer map and the static volume is a 3D mask covering both kidneys.
Coregistration is performed by brute force optimization using a mutual information metric.
Setup#
import numpy as np
import vreg
import vreg.plot as plt
Get data#
# Static oblique volumes
lk = vreg.fetch('left_kidney')
rk = vreg.fetch('right_kidney')
# Moving volume
mtr = vreg.fetch('MTR')
# Geometrical reference
dixon = vreg.fetch('Dixon_water')
Format data#
Create a mask containing both kidneys (bk) with the geometry of the complete DIXON series
bk = lk.slice_like(dixon).add(rk)
Bounding box#
Extract a bounding box to reduce the size of the volume. This is not necessary but it speeds up the calculation a little as the volume is smaller.
bk = bk.bounding_box()
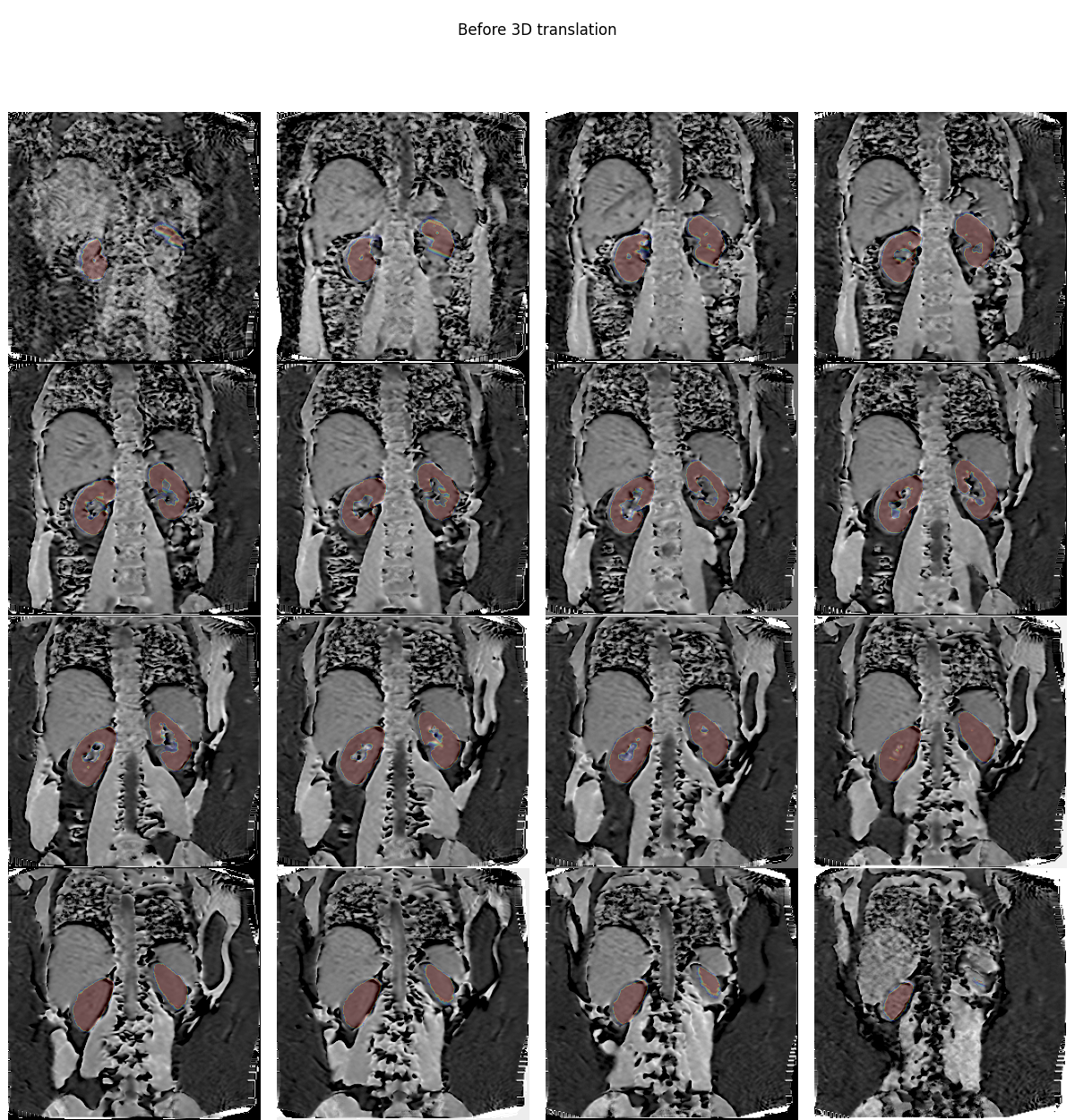
Overlay data before registration#
If we overlay the mask on the volume, we clearly see the misalignment due to different breath holding positions:
plt.overlay_2d(mtr, bk, title='Before 3D translation',
vmin=np.percentile(mtr.values, 10),
vmax=np.percentile(mtr.values, 99))
Coregister#
We are coregistering using a 3D translation in the reference frame of the moving volume. We are using a brute force optimization which is slow but robust. We allow for translations between [-20, 20] mm in-slice, and [-5, 5] mm through-slice, in steps of 2mm.
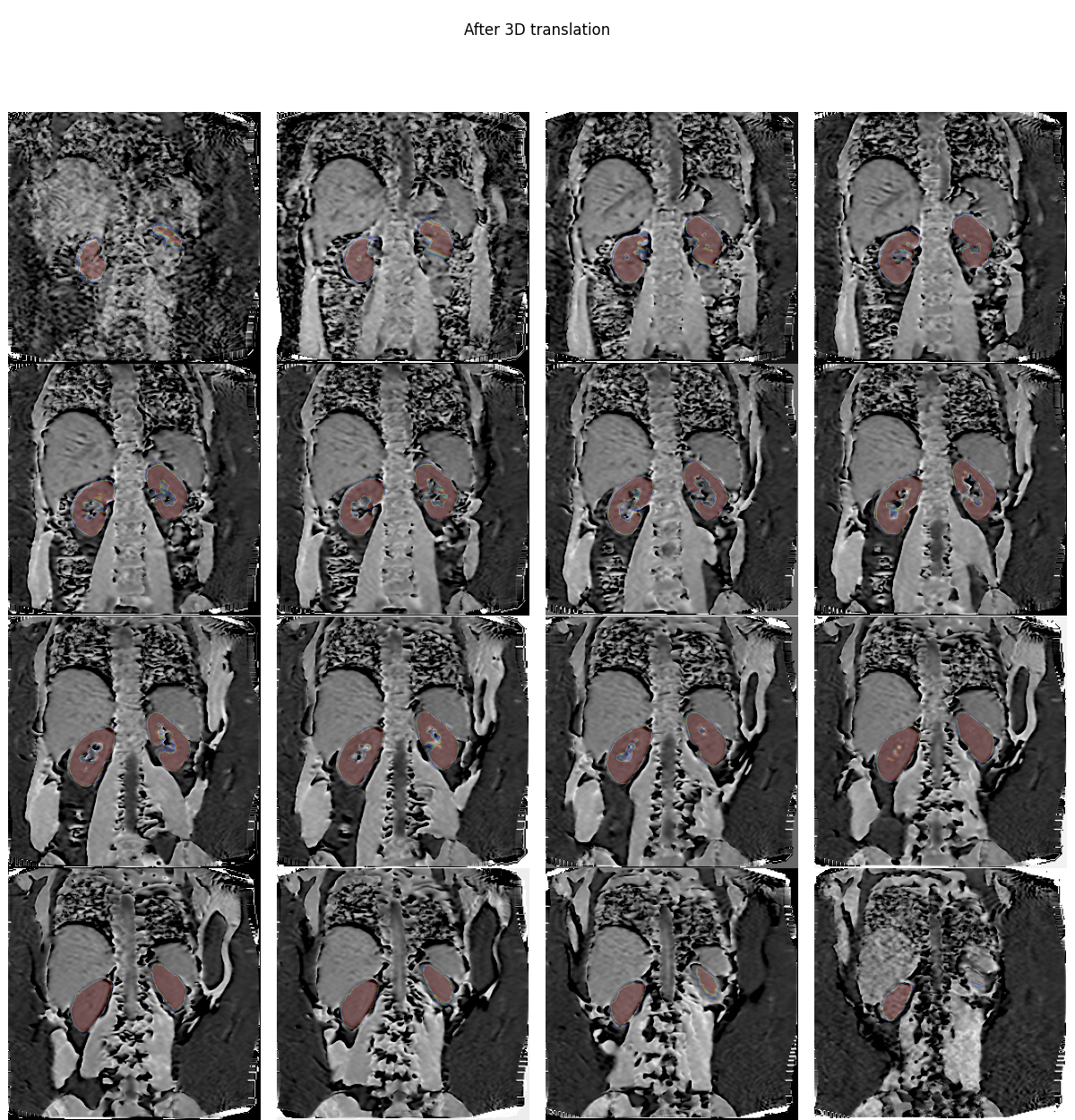
Overlay data after registration#
If we overlay the mask on the new volume, we can see that the misalignment is significantly reduced:
plt.overlay_2d(mtr, bk, title='After 3D translation',
vmin=np.percentile(mtr.values, 10),
vmax=np.percentile(mtr.values, 99))
Total running time of the script: (4 minutes 10.542 seconds)